Missing Data? Impute it.

Uh oh.
A common problem with datasets is missing values in the the rows. It’s a problem that can result from poor data collection, either through inconsistent methodologies, or subsequent changes to protocols and databases during the collection phase.
One tactic to remedy the poor quality data, is to impute (i.e., replace) the missing data points with a quantified measurement. It goes without saying, that this can be risky. But, it can also be a relatively safe practice under certain conditions. If the dataset is large, and the missing data is relatively small, we can interpolate average numerical values from the existing data. In theory, this will allow us to use the rows containing missing data without skewing our results too harshly.
Today, we’ll do exactly that using a Python program I wrote.
Imports
We’ll be using Pandas and TKinter. Pandas for the analytics, and TKinter to generate an easy GUI interface to find your file.
import pandas as pd
import tkinter as tk
from tkinter import filedialog, messagebox
Class
class MissingDataImputer(object):
Selecting Data Files with a tkinter GUI
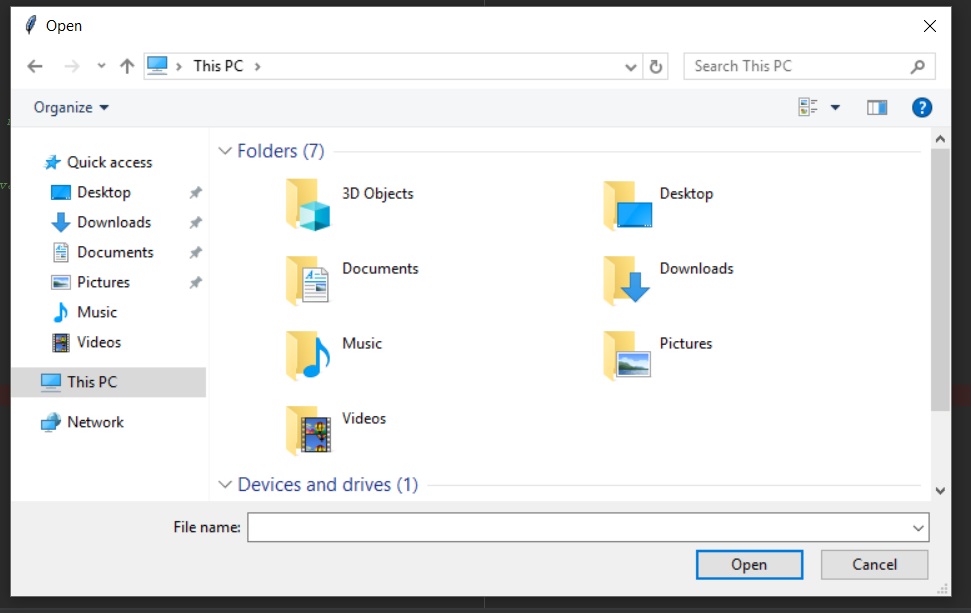
A file selector is a great way to develop an interface for users. In my experience, it’s much easier for users to simply select the file they want, rather than find the path and enter it into a command line interface. Tkinter is a great tool to accomplish this quickly, as I have constructed below.
#Allow User to Select File via tkinter, returns file_path
def getFilePath(self):
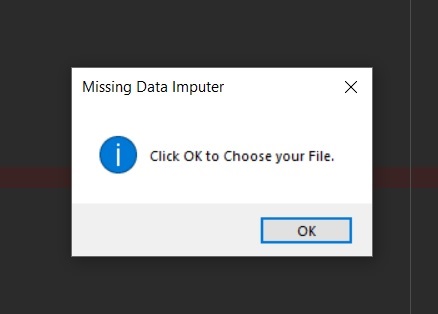
"""Get Name and Location of User's CSV file
Args:
None
Returns:
File Path of Target CSV.
"""
root = tk.Tk()
messagebox.showinfo("Missing Data Imputer", "Click OK to Choose your File.")
root.withdraw()
file_path = filedialog.askopenfilename()
return file_path
Reading Files
Next, we want to load the file we have specified, and store it for further processing, we’ll do this with get_file.
def get_file(self, filename):
""" Extract csv file contents, sep on semi-colon
Args:
filename: Path to target CSV
Returns:
raw data of csv file
"""
raw = pd.read_csv(filename, sep=';')
return raw
Raw Data to Pandas Dataframe
We have the raw data, now let’s construct a DataFrame for easy manipulation via make_dataframe outlined below.
#Convert Raw File to DataFrame
def make_dataframe(self, filecontents):
""" Create Pandas DataFrame of raw file contents
Args:
filecontents: Raw Contents
Returns:
Dataframe of csv file
"""
dataframe = pd.DataFrame(filecontents)
return dataframe
Begin Investigation
Our data is in place, and in a form we can use! Let’s get started with our goal, identifying missing data!
We will search and count the number of missing row indices using list comprehension to iterate over each column. If no missing data is found, we’ll simply print the message to the user and end the program with our worried minds sated.
def check_integrity(self, input_df):
""" Check if values missing, generate list of cols with NaN indices
Args:
input_df - Dataframe
Returns:
List containing column names containing missing data
"""
if df.isnull().values.any():
print("\nDetected Missing Data\nAffected Columns:")
affected_cols = [col for col in input_df.columns if input_df[col].isnull().any()]
affected_rows = df.isnull().sum()
missing_list = []
for each_col in affected_cols:
missing_list.append(each_col)
print(missing_list)
print("\nCounts")
print(affected_rows)
print("\n")
return missing_list
else:
pass
print("\nNo Missing Data Was Detected.")
Missing Data Detected, What Now?
If we identified missing data, we’ll use the method impute() to rectify the problem. We’ll pass our dataframe and list of missing values into this method and use our interpolate() function to impute an interpolated value in place of the missing indices.
def impute(self, input_df, missing):
""" Imputes missing values of detected cols with interpolation methodology
Args:
input_df : dataframe
missing : columns labels associated with missing observations
Returns:
dataframe with interpolated values
"""
for each in missing:
input_df[each] = input_df[each].interpolate()
return input_df
We can then run check_integrity() on our cleaned dataframe to verify that no missing values are present.
Finally, we’ll send our results to our original file.
Printing Results to File
def fileOutput(self, df, file):
""" Replaces file contents with corrected dataframe
Args:
df - dataframe
file - filepath/filename
:return:
None
"""
df.to_csv(file, sep=',', encoding='utf-8')
It’s important to note that this obviously won’t work for binary choices, booleans, strings, or other non-numeric row values. We could still handle them, but we would need different processes and a new post!
Run It
import pandas as pd
import tkinter as tk
from tkinter import filedialog, messagebox
# Load File
class MissingDataImputer(object):
# Allow User to Select File via tkinter, returns file_path
def getFilePath(self):
""" Get Name and Location of User's CSV file
Args:
None
Returns:
File Path of Target CSV.
"""
root = tk.Tk()
messagebox.showinfo("Missing Data Imputer", "Click OK to Choose your File.")
root.withdraw()
file_path = filedialog.askopenfilename()
return file_path
def get_file(self, filename):
""" Extract csv file contents, sep on semi-colon
Args:
filename: Path to target CSV
Returns:
raw data of csv file
"""
raw = pd.read_csv(filename)
return raw
# Convert Raw File to DataFrame
def make_dataframe(self, filecontents):
""" Create Pandas DataFrame of raw file contents
Args:
filecontents: Raw Contents
Returns:
Dataframe of csv file
"""
dataframe = pd.DataFrame(filecontents)
print(dataframe)
return dataframe
def check_integrity(self, input_df):
""" Check if values missing, generate list of cols with NaN indices
Args:
input_df - Dataframe
Returns:
List containing column names containing missing data
"""
if df.isnull().values.any():
print("\nDetected Missing Data\nAffected Columns:")
affected_cols = [col for col in input_df.columns if input_df[col].isnull().any()]
affected_rows = df.isnull().sum()
missing_list = []
for each_col in affected_cols:
missing_list.append(each_col)
print(missing_list)
print("\nCounts")
print(affected_rows)
print("\n")
return missing_list
else:
pass
print("\nNo Missing Data Was Detected.")
def impute(self, input_df, missing):
""" Imputes missing values of detected cols with interpolation methodology
Args:
input_df : dataframe
missing : columns labels associated with missing observations
Returns:
dataframe with interpolated values
"""
for each in missing:
input_df[each] = input_df[each].interpolate()
print(input_df)
return input_df
def fileOutput(self, df, file):
""" Replaces file contents with corrected dataframe
Args:
df - dataframe
file - filepath/filename
:return:
None
"""
df.to_csv(file, sep=',', encoding='utf-8')
if __name__ == "__main__":
run = MissingDataImputer()
your_file_path = run.getFilePath()
file = run.get_file(your_file_path)
df = run.make_dataframe(file)
missing_list = run.check_integrity(df)
clean_df = run.impute(df, missing_list)
run.check_integrity(clean_df)
run.fileOutput(clean_df, your_file_path)
